Fecal Contamination and Microbial Source Tracking

Since 2015, the Environmental Chemistry Lab has been involved with a variety of research projects aimed at monitoring and improving the water quality of Michigan streams and beaches using molecular methods for pathogen detection. In conjunction with the Michigan Department of Environmental Quality, the US Environmental Protection Agency, and various labs across Michigan, we were part of the development and implementation of a qPCR based test method that can provide same day test results for E. coli. Our lab’s data was part of the EPA’s validation study for Method C:
- Sivaganesan, M., Aw, T. G., Briggs, S., Dreelin, E., Aslan, A., Dorevitch, S., Shrestha, A., Isaacs, N., Kinzelman, J., Kleinheinz, G., Noble, R., Rediske, R., Scull, B., Rosenberg, S., Weberman, B., Sivy, T., Southwell, B., Siefring, S., Oshima, K., & Haugland, R. (2019). Standardized data quality acceptance criteria for a rapid Escherichia coli qPCR method (draft method C) for water quality monitoring at recreational beaches. Water Research, 156, 456–464. https://doi.org/10.1016/j.watres.2019.03.011
- Aw, T. Sivaganesan, M., Briggs, S., Dreelin, E., Aslan, A., Dorevitch, S., Shrestha, A., Isaacs, N., Kinzelman, J., Kleinheinz, G., Noble, R., Rediske, R., Scull, B., Rosenberg, S., Weberman, B., Sivy, T., Southwell, B., Siefring, S., Oshima, K., & Haugland, R. (2019). Evaluation of multiple laboratory performance and variability in analysis of recreational freshwaters by a rapid Escherichia coli qPCR method (Draft Method C). Water Research, 156, 465–474. https://doi.org/10.1016/j.watres.2019.03.014
Our lab also led the effort to validate an excel based workbook to standardize the calibration and data reporting for Method C, a comparison that showed the Michigan qPCR Network of labs was able to produce data of similar quality as the EPA Research Lab, and a validity assessment of Michigan’s proposed qPCR beach standard:
- Lane, M.J., McNair, J.N., Rediske, R., Briggs, S., Sivaganesan, M. and Haugland, R., 2020. Simplified Analysis of Measurement Data from A Rapid E. coli qPCR Method (EPA Draft Method C) Using A Standardized Excel Workbook. Water, 12(3), p.775. https://doi.org/10.3390/w12030775
- Lane, M.J., Rediske, R., McNair, J.N., Briggs, S., Rhodes, G., Dreelin, E., Sivy, T., Flood, M., Scull, B., Szlag, D. and Southwell, B., 2020. A comparison of E. coli concentration estimates quantified by the EPA and a Michigan laboratory network using EPA Draft Method C. Journal of Microbiological Methods, 179, p.106086. https://doi.org/10.1016/j.mimet.2020.106086
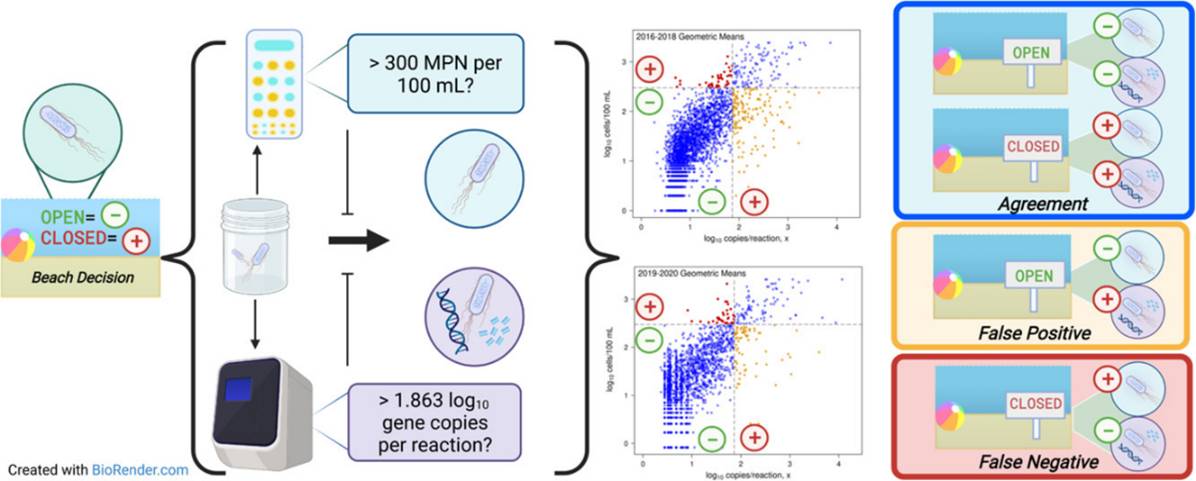
- McNair, J. N., Lane, M. J., Hart, J. J., Porter, A. M., Briggs, S., Southwell, B., ... & Rediske, R. R. 2022. Validity Assessment of Michigan's Proposed qPCR Threshold Value for Rapid Water-Quality Monitoring of E. coli Contamination. Water Research, 119235. https://doi.org/10.1016/j.watres.2022.119235
We have been monitoring west Michigan beaches since 2001 and all our data are posted in BeachGuard.
In addition to E. coli monitoring, we are placing increased emphasis on DNA based methods to determine the sources of fecal pollution in watersheds. This technique, termed Microbial Source Tracking (MST), uses genetic markers liked to host specific gut microbes and can be employed to detect and quantify fecal pollution originating from several sources, including human, bovine, porcine, avian, dog, etc. We part of a 14 lab nationwide validation of the novel NIST Standard Reference Material plasmid, SRM® 2917, for 12 MST markers:
- Sivaganesan, M., Willis, J. R., Karim, M., Babatola, A., Catoe, D., Boehm, A. B., …, Lane, M., Rediske, R., ... & Shanks, O. C. 2022. Interlaboratory Performance and Quantitative PCR Data Acceptance Metrics for NIST SRM® 2917. Water Research, 119162. https://doi.org/10.1016/j.watres.2022.119162
We also seek to link landscape variables with pathogen indicators to better understand and model their interactions and help guide the development of targeted remediation strategies for nonpoint source pollution and the development of Best Management Practices.
Representative projects include:
- E. coli and MST Assessment for Crockery Creek and Sand Creek. Ottawa Conservation
- District.
- E. coli and MST Assessment for the Pigeon River. Ottawa Conservation District.
- E. coli MST Assessment for Bass River and Deer Creek. Ottawa Conservation District.
- E. coli MST Assessment for the Paw Paw River and Black River. Two Rivers Coalition.
- Influence of Tributaries on Beach Water Quality. Michigan Department of the Environment, Great Lakes, and Energy.
